Last updated on March 19, 2024 pm
重命名R1、R2
重命名前
├── SRR12391722
重命名后
SRX8890106
进行定量
Running cellranger count ; cellranger 安装
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 #!/bin/bash export PATH=/opt/cellranger/cellranger-6.1.2:$PATH mkdir $work cd $work for sample in ${data} /*;do echo $sample ${sample##*/} id =$sample_res \$db \$sample \$sample_res \done
附加:scVelo 细胞轨迹
安装依赖
cellranger
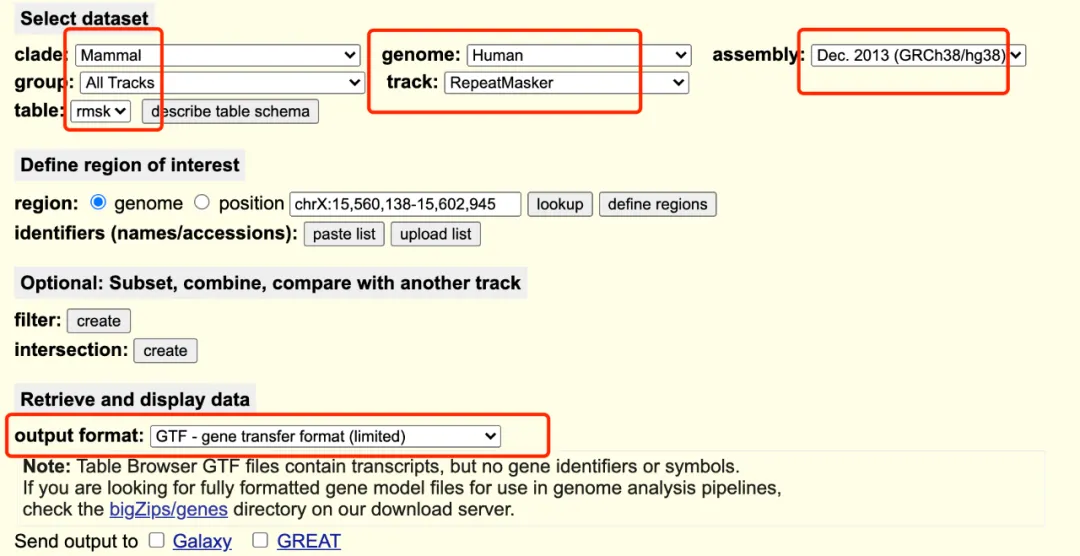
GRCh38_rmsk.gtf.gz:https://genome.ucsc.edu/cgi-bin/hgTables
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 cd /opt/cellrangerexport PATH=/opt/cellranger/cellranger-6.1.2:$PATH id =tiny
velocyto
1 2 3 4 5 6 7 conda create -n velocyto -c conda-forge python=3.7 -yhelp
scVelo
1 2 3 4 5 6 7 8 9 10 11 conda install -c conda-forge widgetsnbextension -yenable --py widgetsnbextension
准备1:运行 cellranger
data
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 #!/bin/bash export PATH=/opt/cellranger/cellranger-6.1.2:$PATH mkdir $work cd $work for sample in ${data} /*;do echo $sample ${sample##*/} id =$sample_res \$db \$sample \$sample_res \done
准备2:从cellranger得到loom文件
1 2 3 4 5 6 7 8 9 10 11 conda activate velocyto${db} /genes/genes.gtfls -lh $rmsk_gtf $work $cellranger_gtf for sample in ${work} /*;do echo $sample $rmsk_gtf $sample $cellranger_gtf done
准备3:从 Seurat 输出 标注 和 UMAP
1 2 3 4 library( Seurat) ( tidyverse) ( stringr) <- readRDS( "~/upload/zl_liu/data/pca.rds" )
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 f_scVelo_group_by <- function ( df, groupN) { <- list ( ) for ( n in unique( as.character ( df[[ groupN] ] ) ) ) { [[ n] ] <- df[ df[[ groupN] ] == n, ] } } <- function ( dfl, cell.embeddings) { for ( n in names ( dfl) ) { [[ n] ] <- cbind( dfl[[ n] ] , cell.embeddings[ rownames( dfl[[ n] ] ) , ] ) } } <- function ( dfl, grepP= '(?=.{10})([AGCT]{16})(?=-1)' ) { for ( n in names ( dfl) ) { ( dfl[[ n] ] ) <- str_extract( rownames( dfl[[ n] ] ) , grepP) } } <- f_scVelo_group_by( sce[[ c ( 'patient_id' , 'cell_type_fig3' ) ] ] , 'patient_id' ) <- f_scVelo_get_reduction( test, sce@ reductions$ umap@ cell.embeddings) <- f_scVelo_get_reduction( test, sce@ reductions$ pca@ cell.embeddings) <- f_scVelo_str_extract_rowN( test)
1 2 3 4 5 6 7 8 9 10 f_scVelo_label_reduction <- function ( dfl, workdir, groupN, outDir) { = file.path( workdir, outDir, 'velocyto' , 'metadata.csv' ) ( dfl[[ groupN] ] , outDir) } = '/home/jovyan/upload/zl_liu/data/data/res' ( test, work, 'patient1' , 'hPB003' ) ( test, work, 'patient3' , 'hPB004' ) ( test, work, 'patient4' , 'hPB006' ) ( test, work, 'patient5' , 'hPB007' )
运行1: 合并数据
第一步 导入模块
1 2 3 4 5 6 7 import scvelo as scvimport scanpy as scimport numpy as npimport pandas as pdimport seaborn as sns 3 'scvelo' )
第二步 读取数据(时间很长)
1 2 3 4 loomf = '/home/jovyan/upload/zl_liu/data/data/res/hPB003/velocyto/hPB003.loom' False )'/home/jovyan/upload/zl_liu/data/data/res/hPB003/velocyto/metadata.csv' 0 )
第三步 取交集并合并数据
1 2 3 4 5 6 7 8 9 10 11 12 13 tmp = [x for x in (x[7 :23 ] for x in adata.obs.index) if x in meta.index]f'hPB003:{x} x' for x in tmp]]'cell_type_fig3' ]'cell_type_fig3' ] = test'X_pca' ] = np.asarray(meta.iloc[:, 4 :])'X_umap' ] = np.asarray(meta.iloc[:, 2 :4 ])'cell_type_fig3' )'cell_type_fig3' )
运行2: 计算绘图
计算
1 2 3 4 5 6 7 8 9 scv.pp.moments(adata, n_pcs=30 , n_neighbors=30 )8 )'dynamical' )8 )'hPB003.h5ad' )
绘图
1 2 3 from matplotlib.pyplot import rc_contextwith rc_context({'figure.figsize' : (12 , 12 )}):'umap' , color=['cell_type_fig3' ], save = "hPB003 velocity embedding stream.svg" )