2022-09-25-【迁移】cellranger定量:One-Library-Multiple-Flowcells.md 8.1 KB
title: 【迁移】cellranger定量:One Library, Multiple Flowcells urlname: cellranger-ding-liang--One-Library--Multiple-Flowcells date: 2022-09-25 19:42:28 index_img: https://api.limour.top/randomImg?d=2022-09-25 19:42:28
tags: ['cellranger', 'scVelo']
重命名R1、R2
-
重命名前
├── SRR12391722
│ ├── SRR12391722_1.fastq.gz
│ └── SRR12391722_2.fastq.gz
├── SRR12391723
│ ├── SRR12391723_1.fastq.gz
│ └── SRR12391723_2.fastq.gz
├── SRR12391724
│ ├── SRR12391724_1.fastq.gz
│ └── SRR12391724_2.fastq.gz
└── SRR12391725
│├── SRR12391725_1.fastq.gz
│└── SRR12391725_2.fastq.gz
重命名后
SRX8890106
├── SRX8890106_S1_L001_R1_001.fastq.gz
├── SRX8890106_S1_L001_R2_001.fastq.gz
├── SRX8890106_S1_L002_R1_001.fastq.gz
├── SRX8890106_S1_L002_R2_001.fastq.gz
├── SRX8890106_S1_L003_R1_001.fastq.gz
├── SRX8890106_S1_L003_R2_001.fastq.gz
├── SRX8890106_S1_L004_R1_001.fastq.gz
└── SRX8890106_S1_L004_R2_001.fastq.gz
进行定量
Running cellranger count; cellranger 安装
#!/bin/bash
export PATH=/opt/cellranger/cellranger-6.1.2:$PATH
db=/opt/cellranger/refdata-gex-GRCh38-2020-A
data=/home/jovyan/work_st/sce/GSE137829/data
work=/home/jovyan/work_st/sce/GSE137829/res
mkdir $work
cd $work
for sample in ${data}/*;
do
echo $sample
sample_res=${sample##*/}
cellranger count --id=$sample_res \
--localcores=12 \
--transcriptome=$db \
--fastqs=$sample \
--sample=$sample_res \
--expect-cells=5000
done
- nano 103.sh
- chmod +x 103.sh
- ./103.sh
附加:scVelo 细胞轨迹
安装依赖
cellranger
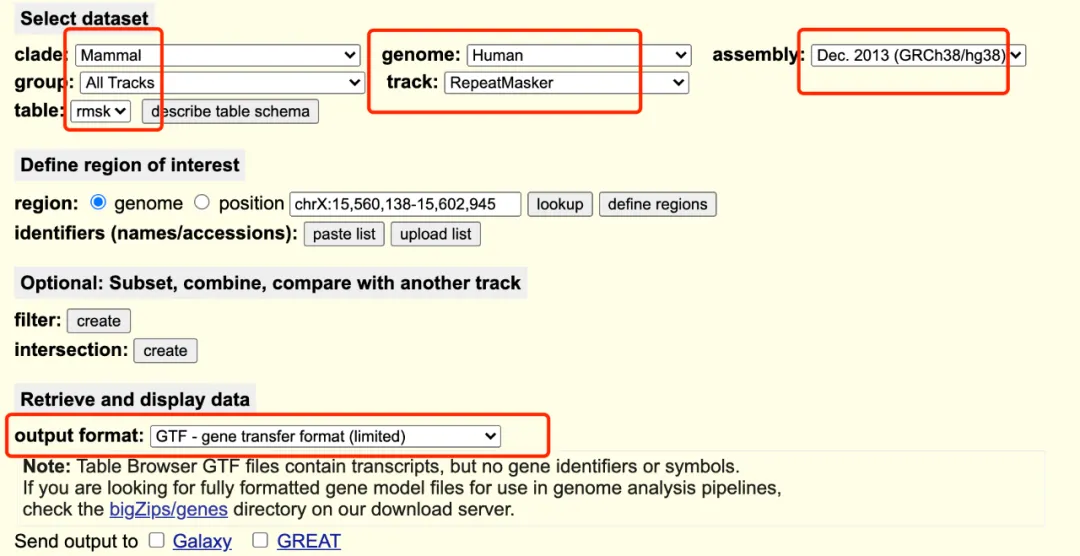
GRCh38_rmsk.gtf.gz:https://genome.ucsc.edu/cgi-bin/hgTables
cd /opt/cellranger
wget <Cell Ranger>
wget <References>
tar -xzvf cellranger-6.1.2.tar.gz
tar -xzvf refdata-gex-GRCh38-2020-A.tar.gz
下载 GRCh38_rmsk.gtf.gz 上传阿里云盘
./aliyunpan
login
d GRCh38_rmsk.gtf.gz -saveto /opt/cellranger
gunzip GRCh38_rmsk.gtf.gz
export PATH=/opt/cellranger/cellranger-6.1.2:$PATH
cellranger sitecheck > sitecheck.txt
cellranger upload xxx@fudan.edu.cn sitecheck.txt
cellranger testrun --id=tiny
cellranger upload xxx@fudan.edu.cn tiny/tiny.mri.tgz
velocyto
conda create -n velocyto -c conda-forge python=3.7 -y
conda activate velocyto
conda install numpy scipy cython numba matplotlib scikit-learn h5py click -y
pip install pysam
pip install velocyto
velocyto --help
conda install -c bioconda samtools=1.15.1 -y
scVelo
conda install -c conda-forge widgetsnbextension -y
jupyter nbextension enable --py widgetsnbextension
# 重启 jupyter
conda create -n scVelo -c conda-forge python=3.7 -y
conda activate scVelo
conda install -c conda-forge scanpy -y
conda install -c conda-forge matplotlib -y
pip install -U scvelo
pip install -U tqdm ipywidgets
conda install -c conda-forge ipykernel -y
python -m ipykernel install --user --name python-scVelo
准备1:运行 cellranger
data
├── hPB003
│ ├── hPB003_S1_L001_R1_001.fastq.gz
│ └── hPB003_S1_L001_R2_001.fastq.gz
├── hPB004
│ ├── hPB004_S1_L001_R1_001.fastq.gz
│ └── hPB004_S1_L001_R2_001.fastq.gz
├── hPB005
│ ├── hPB005_S1_L001_R1_001.fastq.gz
│ └── hPB005_S1_L001_R2_001.fastq.gz
├── hPB006
│ ├── hPB006_S1_L001_R1_001.fastq.gz
│ └── hPB006_S1_L001_R2_001.fastq.gz
└── hPB007
├── hPB007_S1_L001_R1_001.fastq.gz
└── hPB007_S1_L001_R2_001.fastq.gz
#!/bin/bash
export PATH=/opt/cellranger/cellranger-6.1.2:$PATH
db=/opt/cellranger/refdata-gex-GRCh38-2020-A
data=/home/jovyan/upload/zl_liu/data/data/data
work=/home/jovyan/upload/zl_liu/data/data/res
mkdir $work
cd $work
for sample in ${data}/*;
do echo $sample
sample_res=${sample##*/}
cellranger count --id=$sample_res \
--localcores=4 \
--transcriptome=$db \
--fastqs=$sample \
--sample=$sample_res \
--expect-cells=5000
done
准备2:从cellranger得到loom文件
conda activate velocyto
#!/bin/bash
db=/opt/cellranger/refdata-gex-GRCh38-2020-A
work=/home/jovyan/upload/zl_liu/data/data/res
rmsk_gtf=/opt/cellranger/GRCh38_rmsk.gtf # 从genome.ucsc.edu下载
cellranger_gtf=${db}/genes/genes.gtf
ls -lh $rmsk_gtf $work $cellranger_gtf
for sample in ${work}/*;
do echo $sample
velocyto run10x -m $rmsk_gtf $sample $cellranger_gtf
done
准备3:从 Seurat 输出 标注 和 UMAP
library(Seurat)
library(tidyverse)
library(stringr)
sce <- readRDS("~/upload/zl_liu/data/pca.rds")
f_scVelo_group_by <- function(df, groupN){
res <- list()
for(n in unique(as.character(df[[groupN]]))){
res[[n]] <- df[df[[groupN]] == n,]
}
res
}
f_scVelo_get_reduction <- function(dfl, cell.embeddings){
for(n in names(dfl)){
dfl[[n]] <- cbind(dfl[[n]], cell.embeddings[rownames(dfl[[n]]),])
}
dfl
}
f_scVelo_str_extract_rowN <- function(dfl, grepP='(?=.{10})([AGCT]{16})(?=-1)'){
for(n in names(dfl)){
rownames(dfl[[n]]) <- str_extract(rownames(dfl[[n]]), grepP)
}
dfl
}
test <- f_scVelo_group_by(sce[[c('patient_id','cell_type_fig3')]], 'patient_id')
test <- f_scVelo_get_reduction(test, sce@reductions$umap@cell.embeddings)
test <- f_scVelo_get_reduction(test, sce@reductions$pca@cell.embeddings)
test <- f_scVelo_str_extract_rowN(test)
f_scVelo_label_reduction <- function(dfl, workdir, groupN, outDir){
outDir = file.path(workdir, outDir, 'velocyto', 'metadata.csv')
write.csv(dfl[[groupN]], outDir)
}
work='/home/jovyan/upload/zl_liu/data/data/res'
f_scVelo_label_reduction(test, work, 'patient1', 'hPB003')
f_scVelo_label_reduction(test, work, 'patient3', 'hPB004')
f_scVelo_label_reduction(test, work, 'patient4', 'hPB006')
f_scVelo_label_reduction(test, work, 'patient5', 'hPB007')
运行1: 合并数据
第一步 导入模块
import scvelo as scv
import scanpy as sc
import numpy as np
import pandas as pd
import seaborn as sns
scv.settings.verbosity = 3 # show errors(0), warnings(1), info(2), hints(3)
scv.settings.set_figure_params('scvelo') # for beautified visualization
第二步 读取数据(时间很长)
loomf = '/home/jovyan/upload/zl_liu/data/data/res/hPB003/velocyto/hPB003.loom'
adata = scv.read(loomf, cache=False)
metadataf = '/home/jovyan/upload/zl_liu/data/data/res/hPB003/velocyto/metadata.csv'
meta = pd.read_csv(metadataf, index_col=0)
第三步 取交集并合并数据
tmp = [x for x in (x[7:23] for x in adata.obs.index) if x in meta.index]
meta = meta.loc[tmp]
adata = adata[[f'hPB003:{x}x' for x in tmp]]
test = meta['cell_type_fig3']
test.index = adata.obs.index
adata.obs['cell_type_fig3'] = test
adata.obsm['X_pca'] = np.asarray(meta.iloc[:, 4:])
adata.obsm['X_umap'] = np.asarray(meta.iloc[:, 2:4])
sc.pl.pca(adata, color='cell_type_fig3')
sc.pl.umap(adata, color='cell_type_fig3')
运行2: 计算绘图
计算
scv.pp.moments(adata, n_pcs=30, n_neighbors=30)
scv.tl.recover_dynamics(adata, n_jobs=8)
scv.tl.velocity(adata, mode='dynamical')
scv.tl.velocity_graph(adata, n_jobs=8)
adata.write('hPB003.h5ad')
绘图
from matplotlib.pyplot import rc_context
with rc_context({'figure.figsize': (12, 12)}):
scv.pl.velocity_embedding_stream(adata, basis='umap', color=['cell_type_fig3'], save = "hPB003 velocity embedding stream.svg")